Scientists need a thorough knowledge of previous literature to discover new treatments for genetic disorders. Having the right knowledge helps them determine the best gene or protein targets, along with promising drugs for testing. However, since biomedical literature is growing drastically, it contains conflicting information. It makes it difficult for researchers to conduct and complete reviews.
Therefore, to address this, Cole Deisseroth, a graduate student enrolled in the M.D./Ph.D. program and mentored by Drs. Huda Zoghbi and Zhandong Liu at the Jan and Duncan Neurological Research Institute (Duncan NRI) at Texas Children’s Hospital and Baylor College of Medicine, did a study to generate a natural language processing tool. It is called PARMESAN (Parsing Modifiers via Article Annotations).
What does PARMESAN do?
PARMESAN, the new tool, can search for up-to-date information. Moreover, it can be assembled into a central knowledge base and predict the drugs that can correct certain protein imbalances.
The description and capabilities of the tool were published recently in the American Journal of Human Genetics.
Dr Huda Zoghbi said,
PARMESAN offers a wonderful opportunity for scientists to speed up the pace of their research and thus, accelerate drug discovery and development,
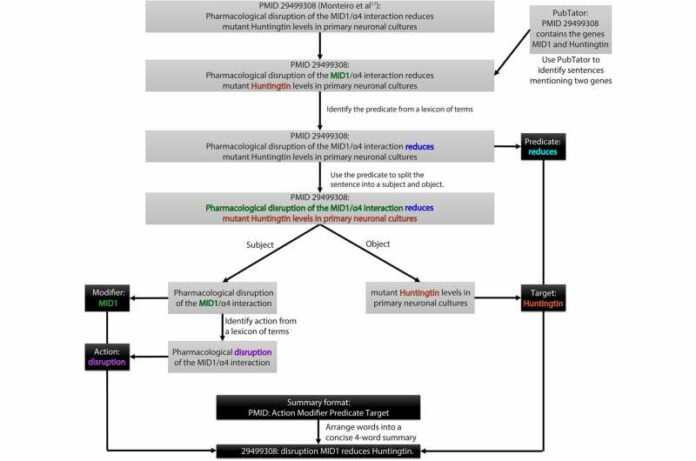
This AI-powered tool scans public biomedical literature databases. It identifies and ranks descriptions of gene-gene and drug-gene regulations. However, PARMESAN stands out because it has the leverage of curated information for predicting undiscovered relationships.
The AI algorithms analyze studies describing the contributions of multiple players in a multistep genetic pathway. After which, a weighted numerical score is assigned to every reported interaction. Consistent and frequent interactions have a higher score. While those with fewer interactions or contradicted have a lower score.
Dr Zhandong Liu said,
The unique feature of PARMESAN is that it not only identifies existing gene-gene or drug-gene interactions based on the available literature but also predicts putative novel drug-gene relationships by assigning an evidence-based score to each prediction,
PARMESAN provides predictions for more than 18,000 target genes currently. Moreover, the benchmark studies suggest that the highest-scoring predictions are more than 95% accurate.
Cole Deisseroth said,
By pinpointing the most promising gene and drug interactions, this tool will allow researchers to identify the most promising drugs at a faster rate and with greater accuracy,